Syntax
Turtle Syntax
We invite you to explore Turtle (Terse RDF Triple Language), a specialized syntax designed for articulating data in the RDF (Resource Description Framework) format. Turtle, while sharing some syntactic similarities with SPARQL (SPARQL Protocol and RDF Query Language), stands out due to its unique application and functionality. It is primarily used for representing and conveying RDF data efficiently, offering a format that is both compact and easily understandable.
Key Elements of Turtle/RDF Syntax:
- IRIs: IRIs are a superset of URLs that are utilized to identify resources, properties, or values, IRIs in Turtle are typically
enclosed in angle brackets, like
<>. Commonly IRIs take the form of URLs when we deal with web resources. - Prefixes: In Turtle, these are employed to abbreviate lengthy IRI namespaces, enhancing the readability.
- Triples: The core of RDF syntax, a triple consists of a subject (the resource or entity being described), a predicate (defining the connection or association between subject and object), and an object.
- Literals: Often used as objects in RDF, literals are enclosed in quotation marks. They represent values such as strings, numbers, and dates.
- Other links and References:
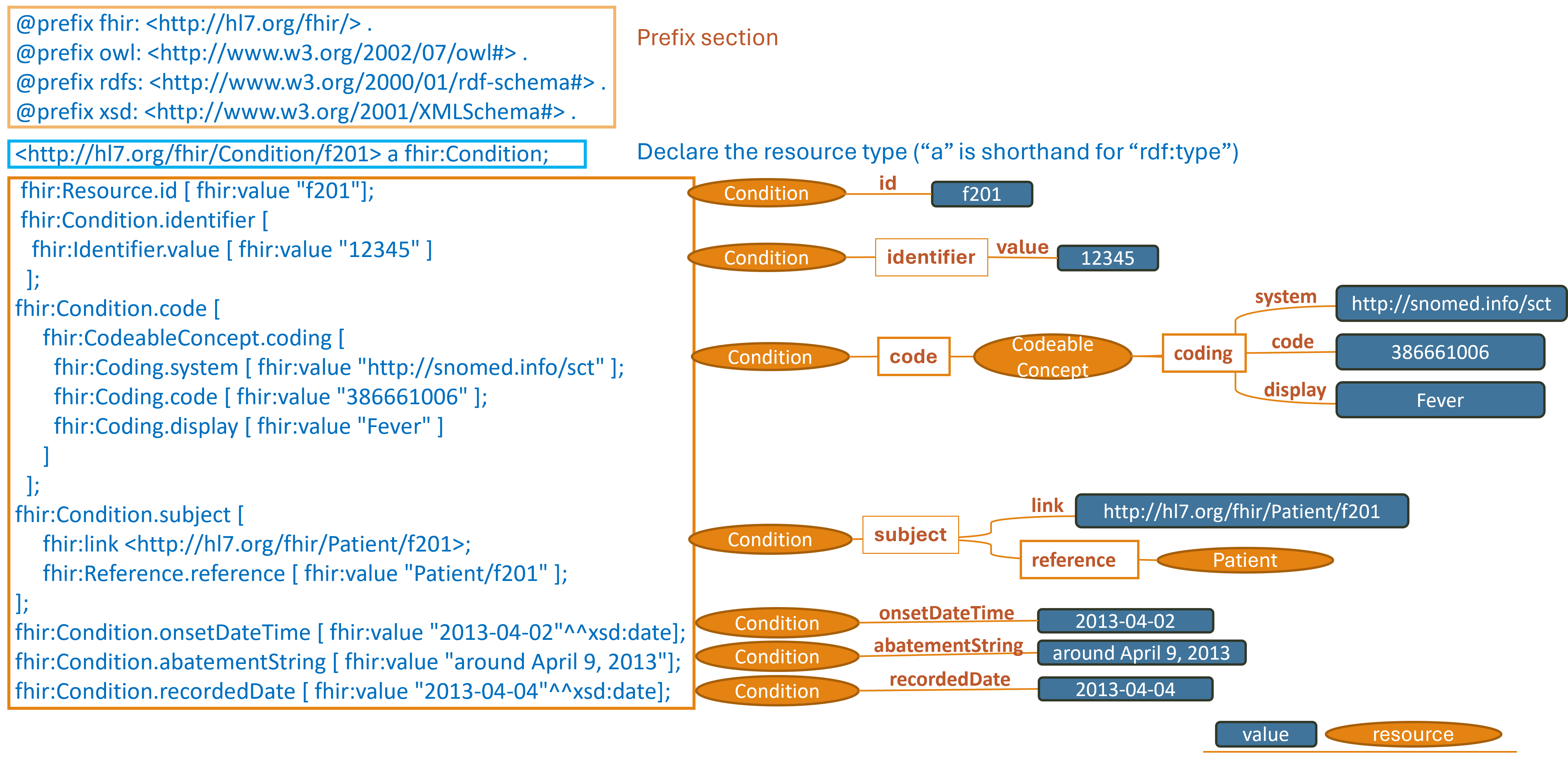
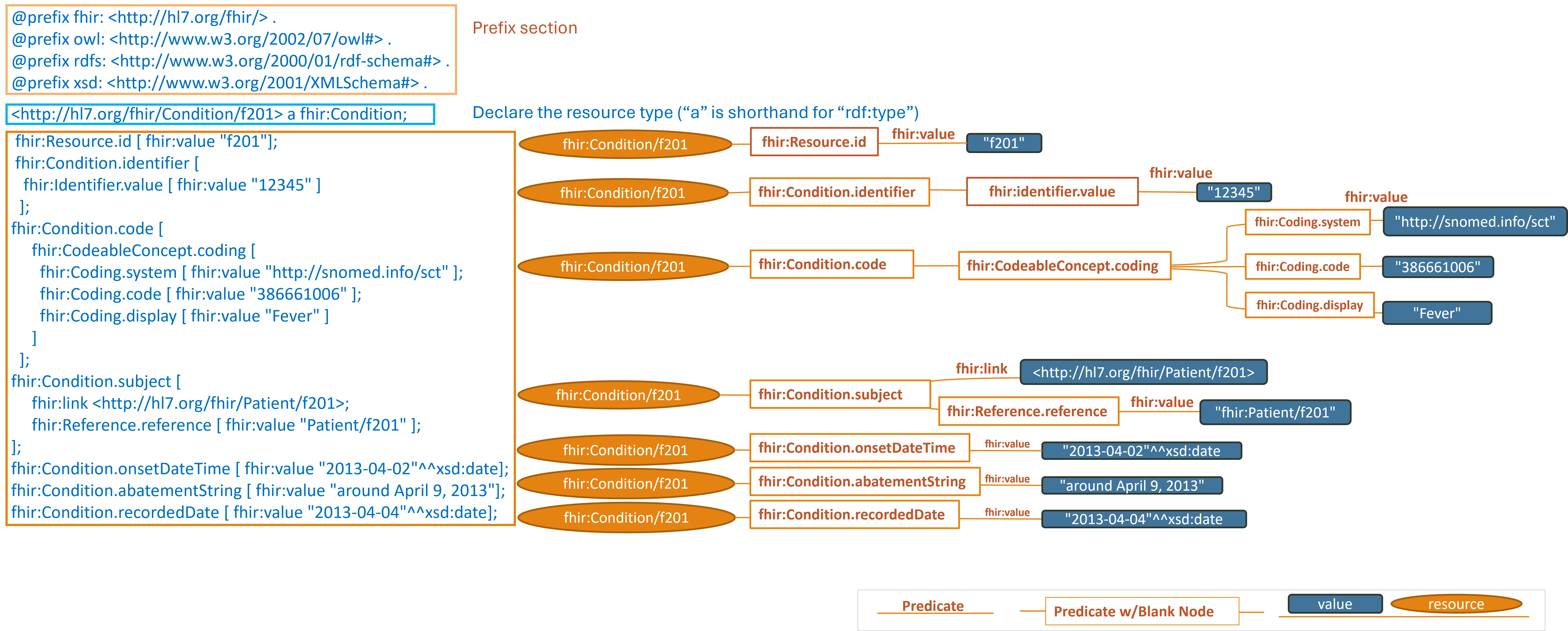
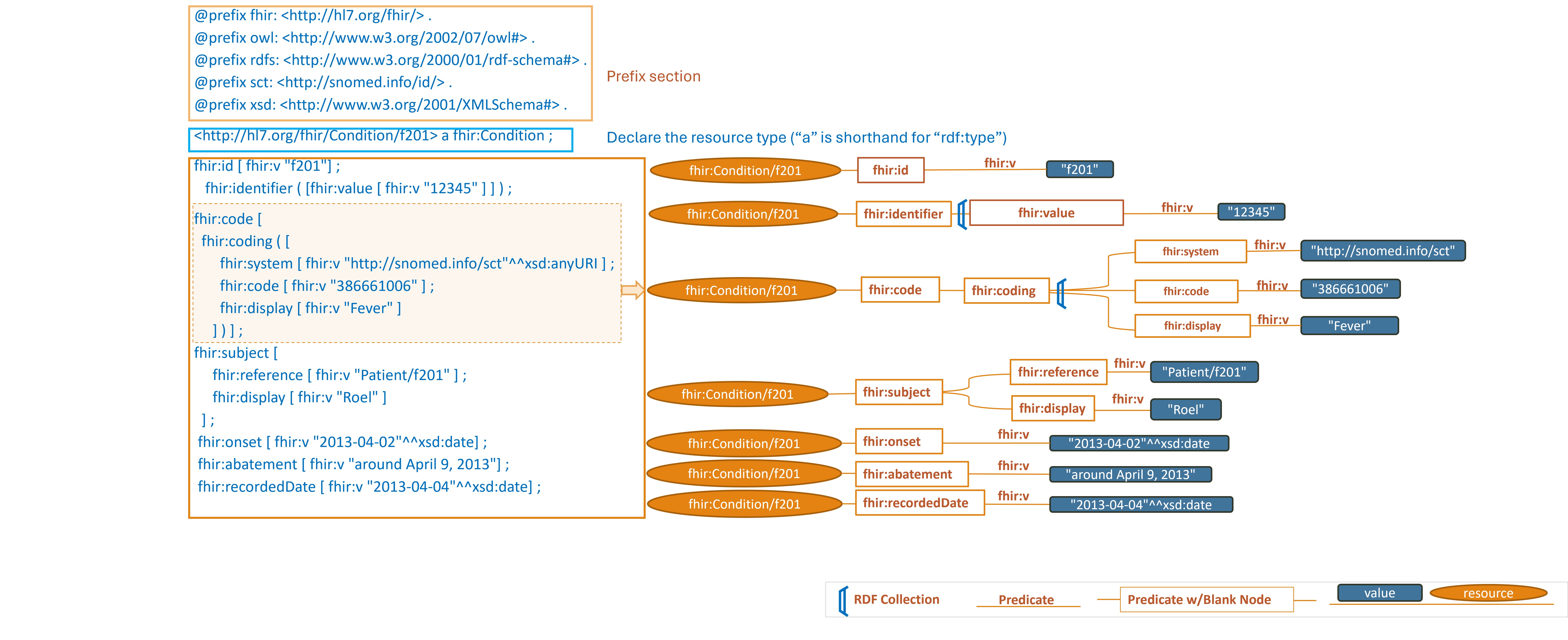
https://hl7.org/fhir/R4/condition-example-f201-fever.ttl2.html
Note: simplified graph does not represent accurate IRIs in FHIR RDF. See other display options below for fully formed IRIs.
This section describes the triple patterns that http://hl7.org/fhir/Condition/f201 has and the
locations where the values of those objects can be found.
Turtle Syntax key points
- In Turtle and RDF,
ais a predicate that is shorthand forrdf:typeor<http://www.w3.org/1999/02/22-rdf-syntax-ns#type>when written as an absolute IRI - Many of the lines end with a semicolon
";"rather than a period. The semicolon in Turtle syntax serves as a separator between predicate-object pairs within a subject block. - When a subject has multiple predicate-object pairs associated with it, using semicolons presents the relationships in a more compact form.
- If there is only one predicate-object pair for a subject, the semicolon can be omitted, and the triple should be ended with
a period
"." - A similar pattern is observed by using commas
","to continue associating objects to the preceding subject and predicate. - The term
fhir:valueappears frequently above. In FHIR,fhir:valueis used to denote the value of a specific property or element within a FHIR resource. It is commonly combined with a series of other property qualifiers to provide additional context or constraints for a value.
Below are some additional line-by-line examples of Turtle Syntax
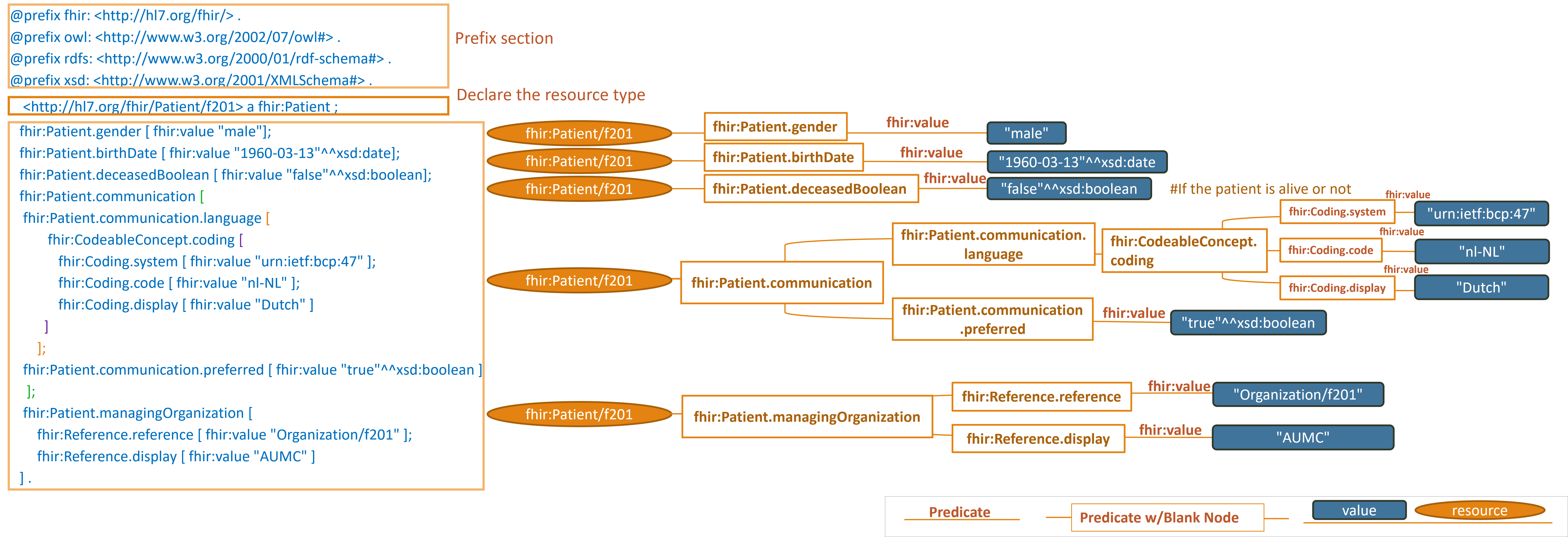
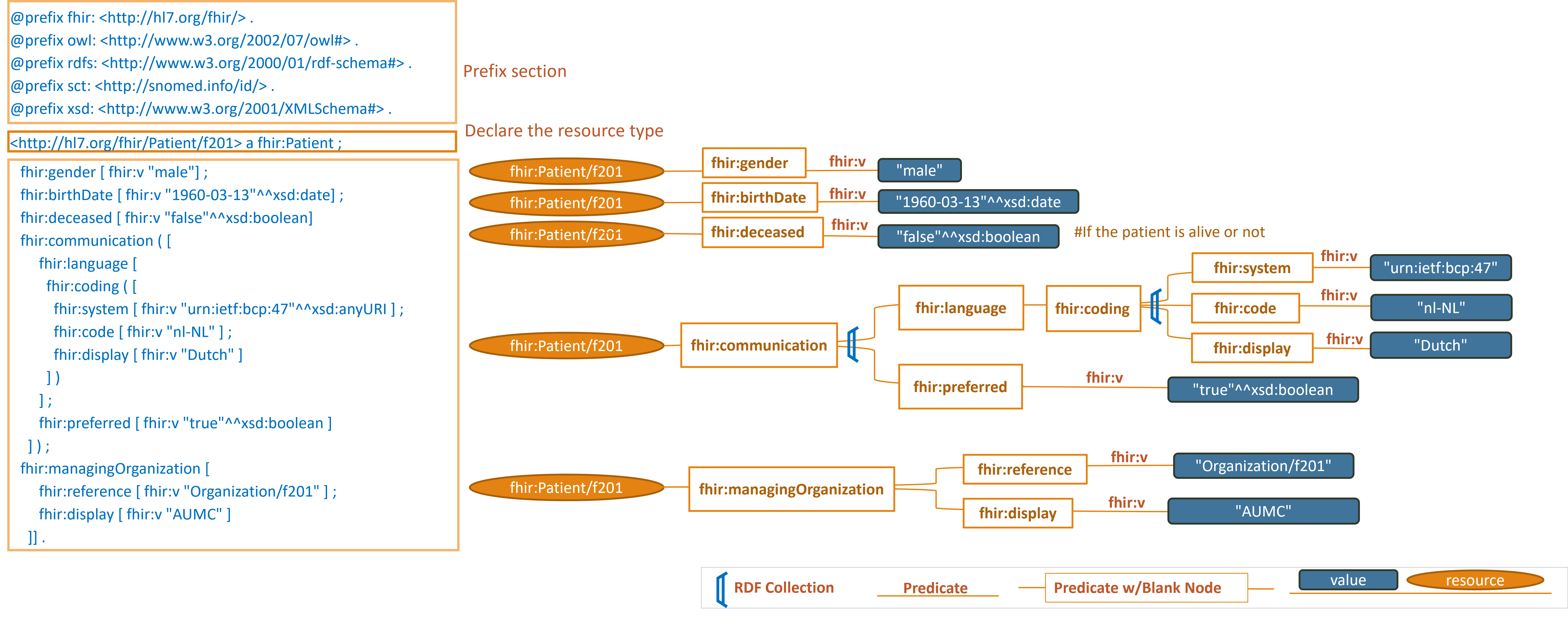
Example 1:
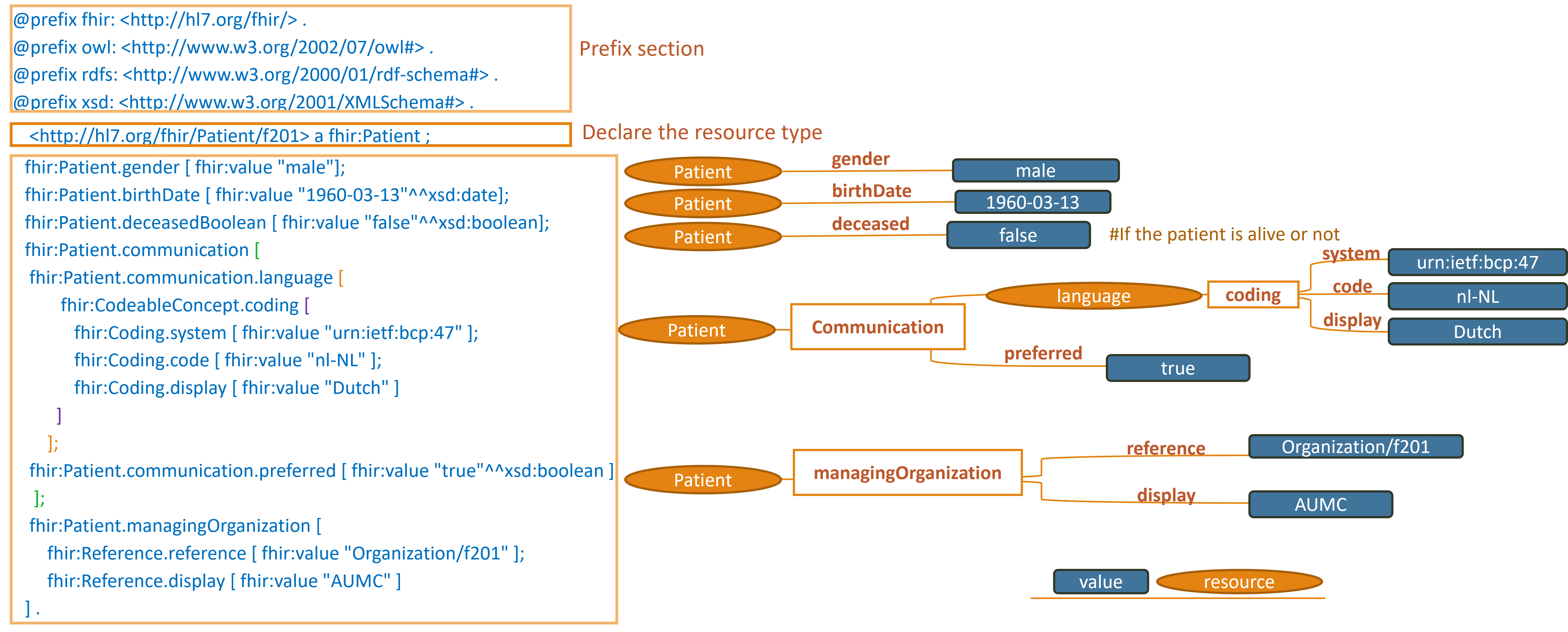
https://hl7.org/fhir/R4/patient-example-f201-roel.ttl.html
Note: simplified graph does not represent accurate IRIs in FHIR RDF. See other display options below for fully formed IRIs.
Example 2:
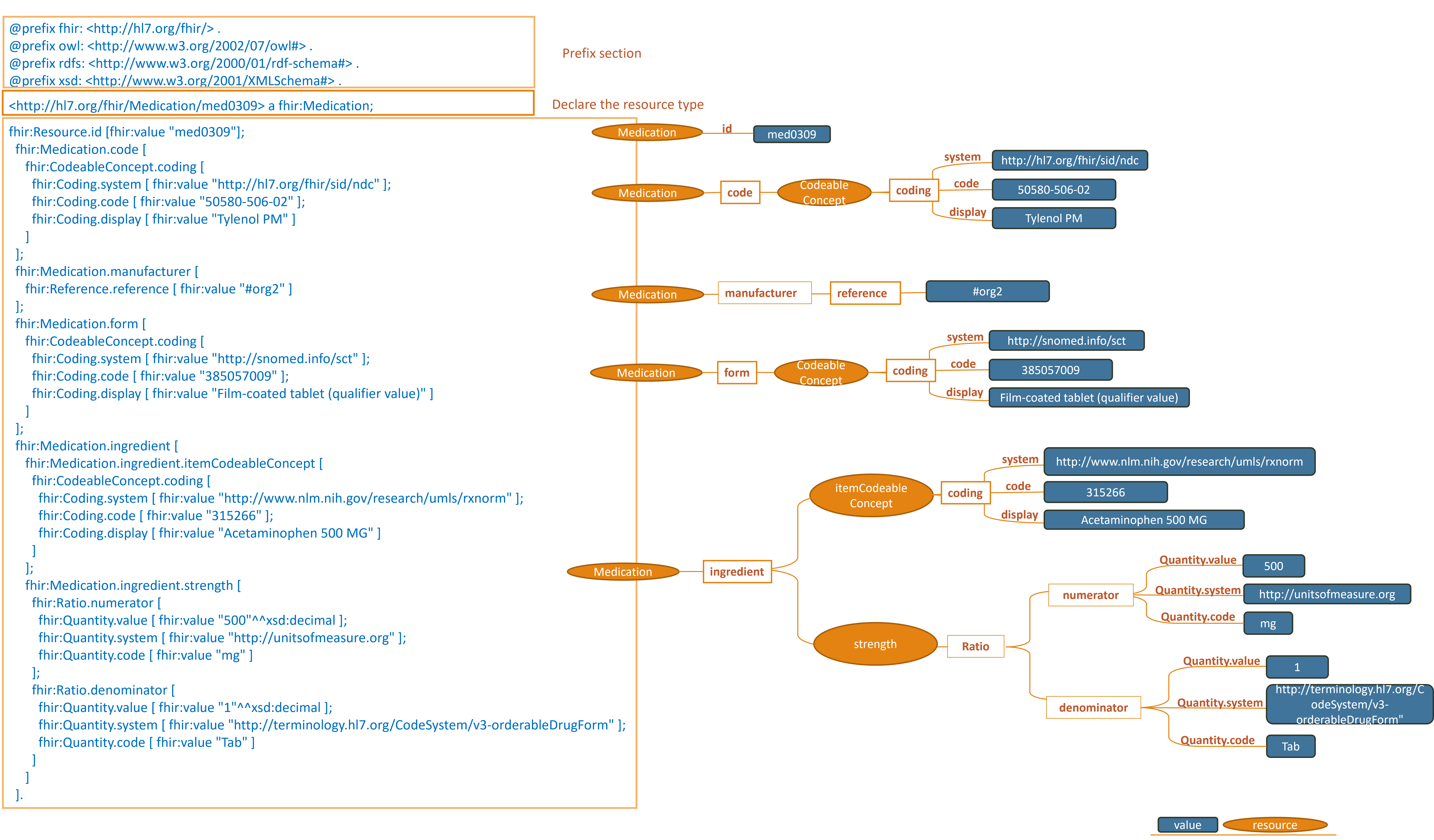
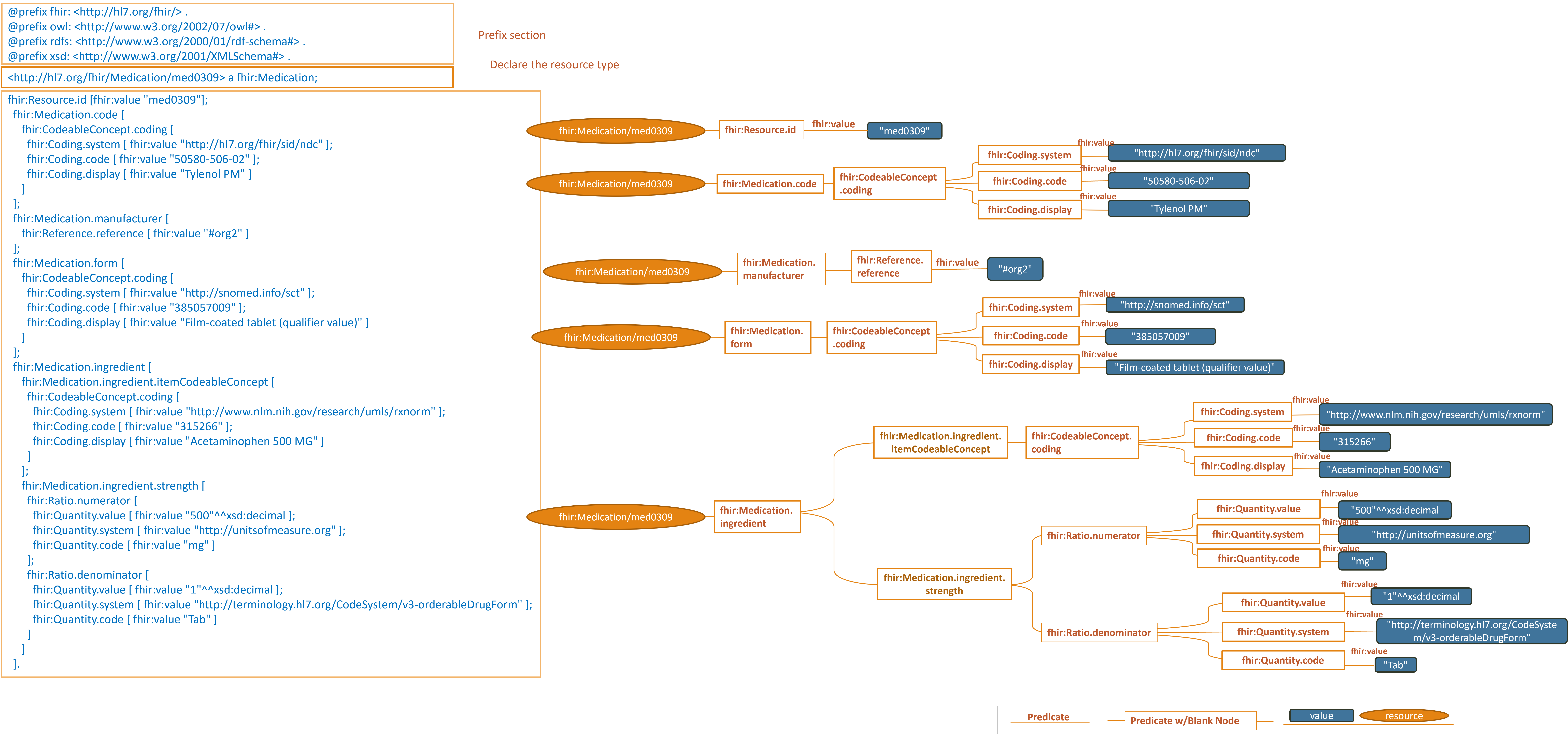
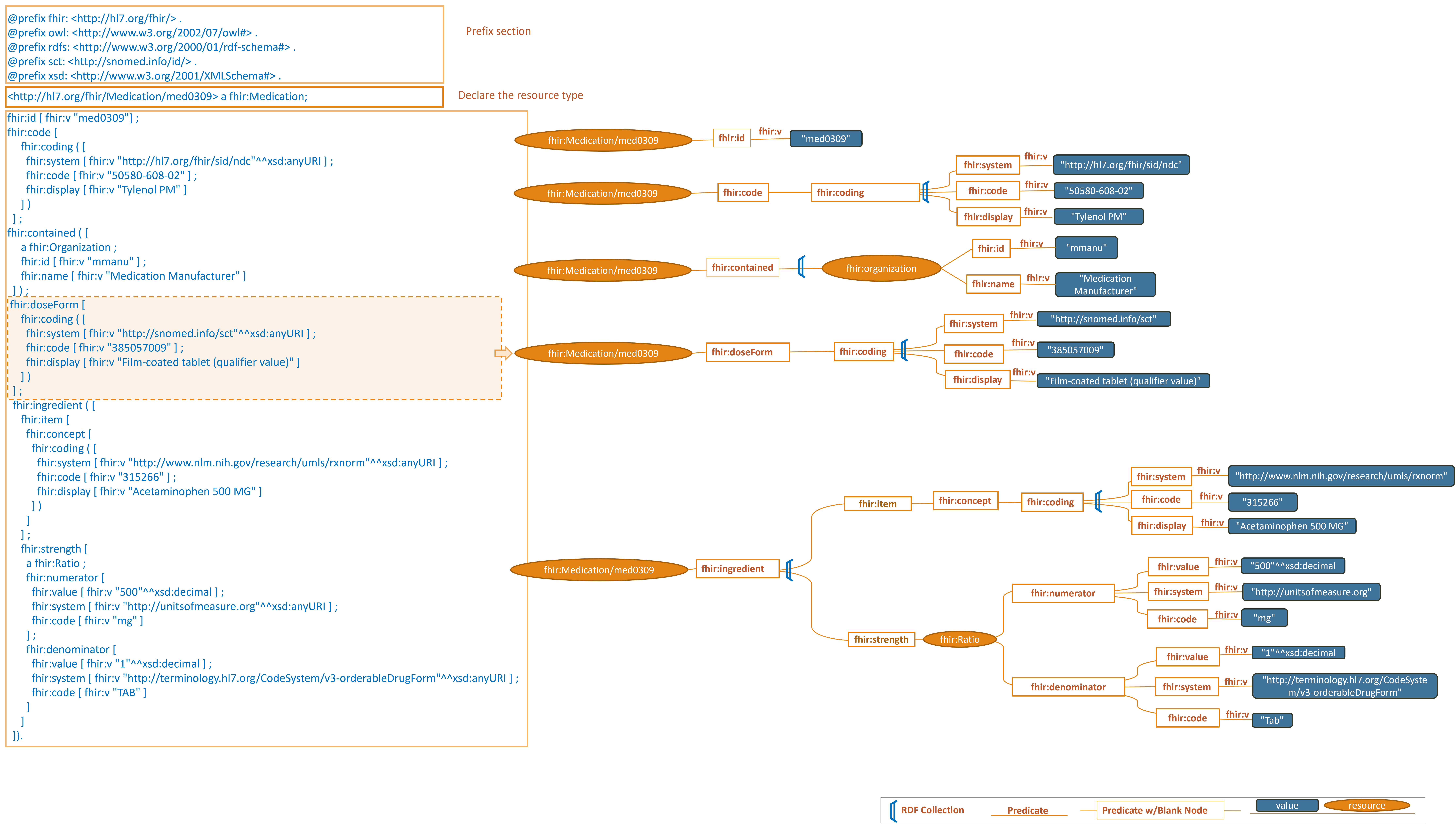
https://hl7.org/fhir/R4/medicationexample0309.ttl.html
Note: simplified graph does not represent accurate IRIs in FHIR RDF. See other display options below for fully formed IRIs.
SPARQL Syntax
Syntax and Structure of SPARQL
Similar to SQL queries, SPARQL queries consist of several clauses, including but not limited to
SELECT, WHERE, OPTIONAL, and FILTER. These clauses work
together to retrieve and filter data from RDF graphs,
allowing for powerful and precise data manipulation.
The SELECT clause details what data to retrieve, while the WHERE clause specifies
the data's patterns and conditions. OPTIONAL allows for the inclusion of data that may not meet all
query conditions, and FILTER refines the results to meet specified criteria.
Similar to Turtle, it's common to begin with a prefix section to simplify writing commonly used IRIs/URLs and enhance readability.
TIP: see more of the SPARQL syntax used in the OMOP VKG Demo Playground
SPARQL Example 1
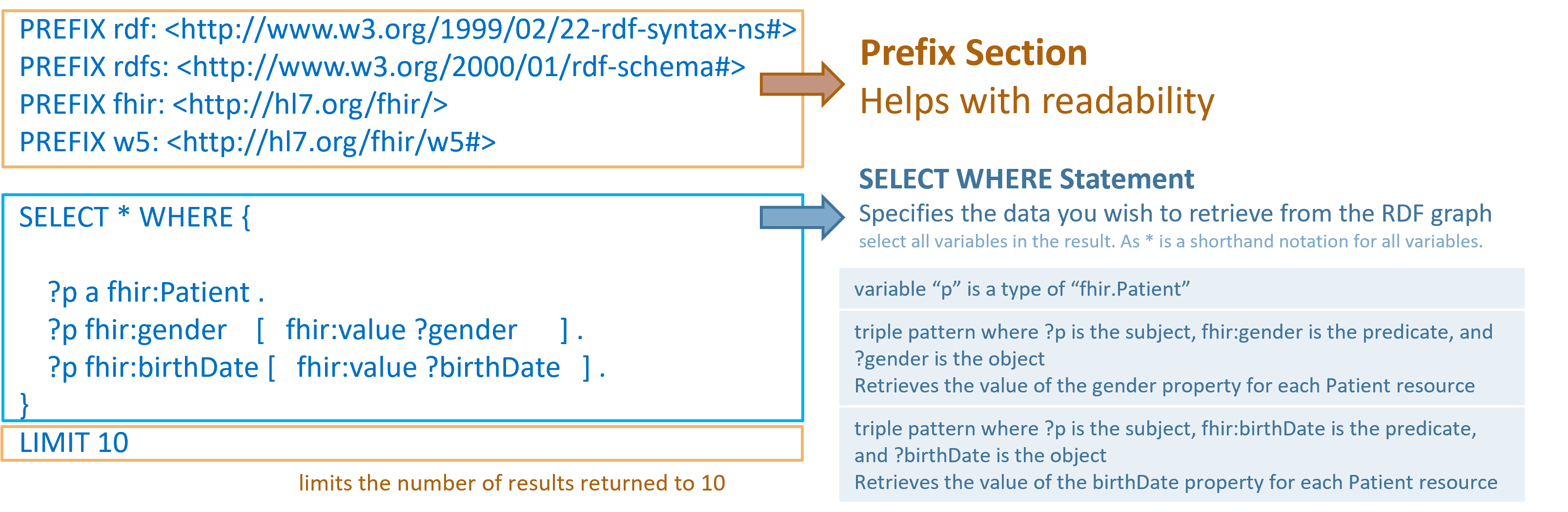
Some triple patterns include square brackets "[]", which denote blank nodes, allowing for the
extraction of values where explicit data may not be available. This is illustrated in the line
?p fhir:gender[fhir:value ?gender], where ?p is a variable representing a patient,
and the blank node provides a flexible way to query associated gender values.
SPARQL Example 2
In many cases, you may want to explore the characteristics of a resource that is the object of another resource.
For
this, the fhir:reference and fhir:link predicate is used to establish the connection.
For example, to examine the characteristics of a patient associated with a particular condition, the query
would
be constructed as follows:
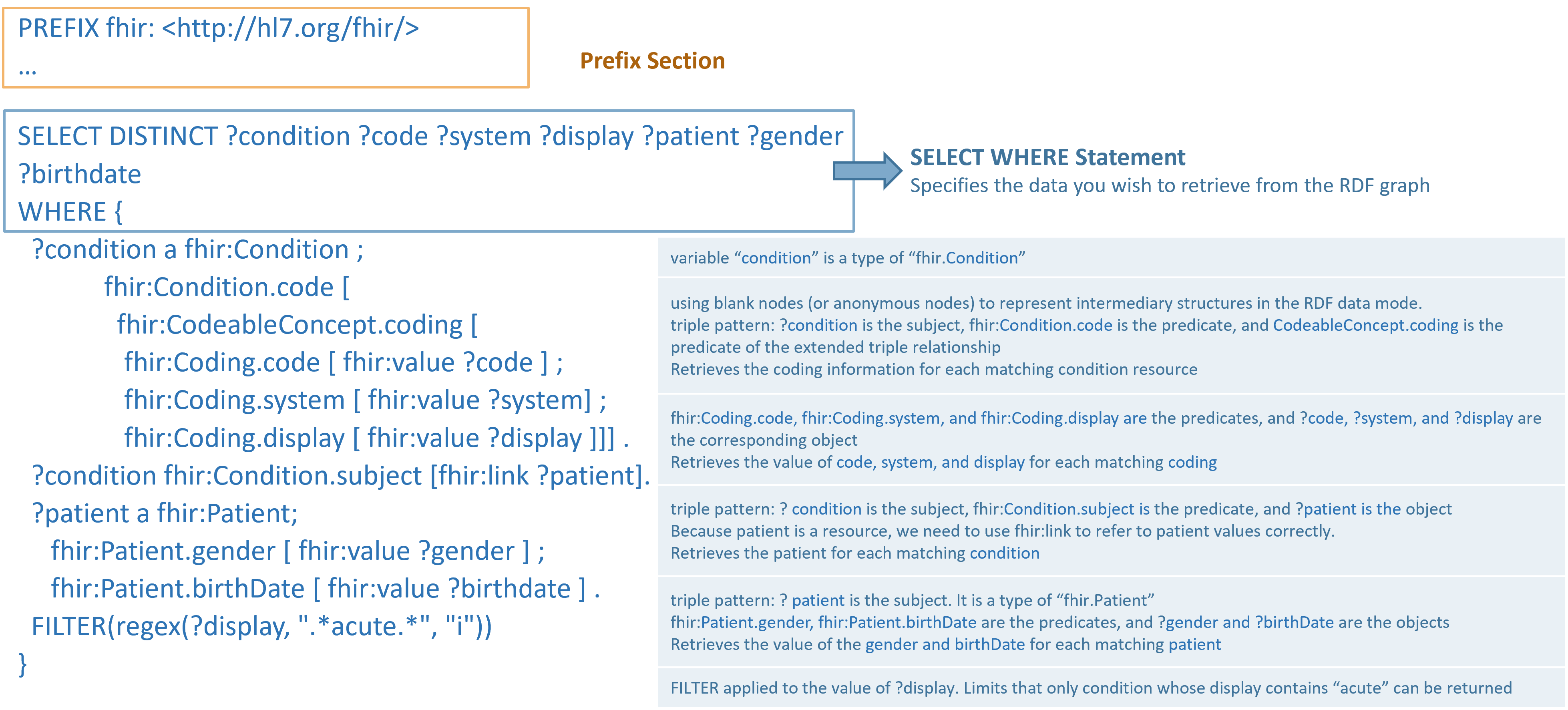
In the example above, semicolons ";" are used at the end of lines within the SPARQL query. A
semicolon indicates that the following predicate-object pairs are associated with the same subject from the previous line.
This allows for the grouping of related information about a single subject neatly. For instance:
In the line ?patient a fhir:Patient;
fhir:Patient.gender [ fhir:value ?gender ] ;
fhir:Patient.birthDate [ fhir:value ?birthdate ] .
This query snippet is seeking to retrieve patient gender and birthdate information where the subject ?patient has
associated values the gender and birthdate properties.
SPARQL's syntax is designed to accommodate complex queries within the semantic web, offering intricate mechanisms for pattern matching, optional values, and condition filtering. Its capabilities are essential for querying and navigating the interconnected data found in RDF triples.
SQL Syntax
SQL, or Structured Query Language, is the most widely adopted language used to query relational databases. It encompasses a range of operations from data querying to database modification and management. While a standard has been declared, many different implementations do not fully cover the full standard and introduce their own idiosyncrasies.
Common Key SQL clauses for querying data include:
SELECT: Specifies the columns to be displayed in the query results.FROM: Indicates the tables from which data is to be retrieved.WHERE: Applies conditions to filter the dataset.JOIN: Combines rows from multiple tables based on a related column.GROUP BY: Aggregates rows with identical values in certain columns.ORDER BY: Sorts the results in either ascending (default) or descending order.LIMIT: Sets a cap on the number of rows to be returned in the result set.
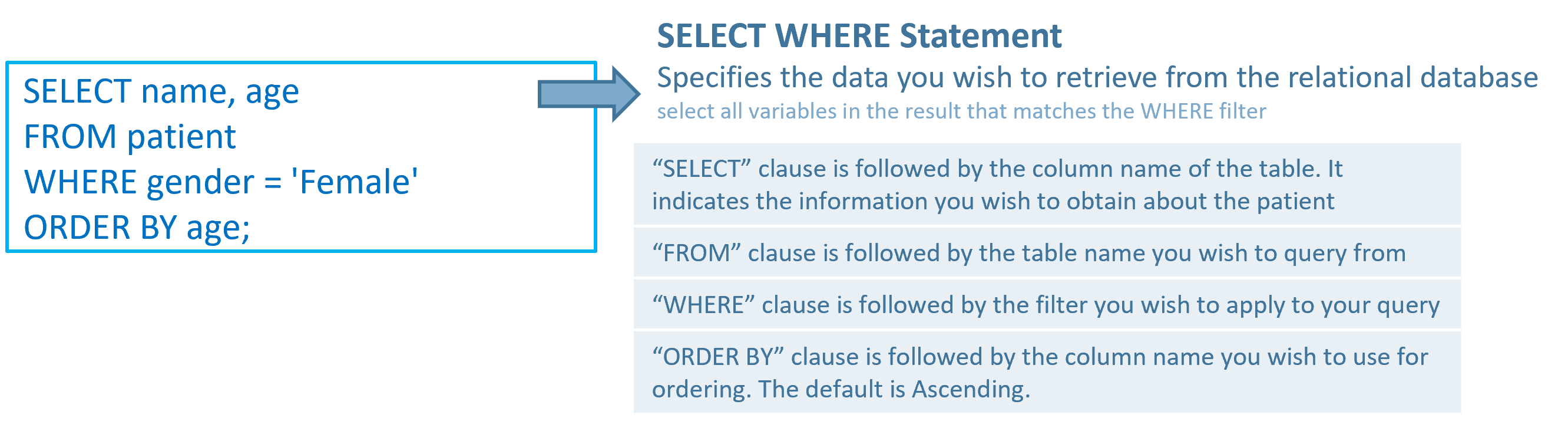
For example, if we want to use SQL to query OMOP CDM to locate the name and age of female patients, we can use the following query.
Healthcare professionals often encounter complex data tasks, such as identifying the most recent patient
observations. For such tasks, if a date record column is present in the database, use the
MAX function to select the latest date, pinpointing the most current observations.

SQL's versatility extends beyond querying. It includes clauses like UPDATE and SET for
modifying data, as well as commands like CREATE TABLE, ALTER TABLE,
DROP TABLE, INSERT INTO, and DELETE for database structure management. Its
powerful and straightforward approach to database manipulation makes SQL indispensable for relational database
management.
Summary
SQL, SPARQL, and Turtle each play a vital role in the healthcare industry, empowering professionals with diverse
capabilities for querying and managing data. SQL's robust transactional commands make it ideal for structured,
table-oriented data management in relational databases.
On the other hand, SPARQL and Turtle shine in the semantic web domain; SPARQL's sophisticated querying enables
intricate searches across complex data relationships, while Turtle's syntax provides a human-readable way to
represent RDF data, making semantic data more accessible.
These languages represent different facets of data handling: SQL for traditional database management, SPARQL
for navigating RDF data within linked data structures, and Turtle for concisely conveying RDF content. Together,
they offer a comprehensive set of tools that address the specific needs of the evolving digital landscape in
healthcare data management.