FHIR and OMOP CDM comparisons
FHIR Structure
As detailed in previous sections, the FHIR framework is constructed using a suite of modular components known as "Resources." These resources form the backbone of the FHIR data standard, each serving as a building block for representing a wide array of healthcare-related data. The utilization of these resources is governed by "Profiles" — customizations of resources that specify which elements are used, and how they are used for different healthcare scenarios. This dual structure of resources and profiles provides an elegant solution to the challenge of data variability in healthcare processes. It ensures universal readability across systems, while allowing applications to introduce additional layers of control and specificity through profiles. FHIR R4 boasts support for 146 distinct resources, encompassing administrative concepts such as patients, providers, organizations, and devices, alongside a broad spectrum of clinical concepts including problems, medications, diagnostics, care plans, and financial matters, among others. This enables healthcare professionals to selectively implement the resources most pertinent to their specific needs.
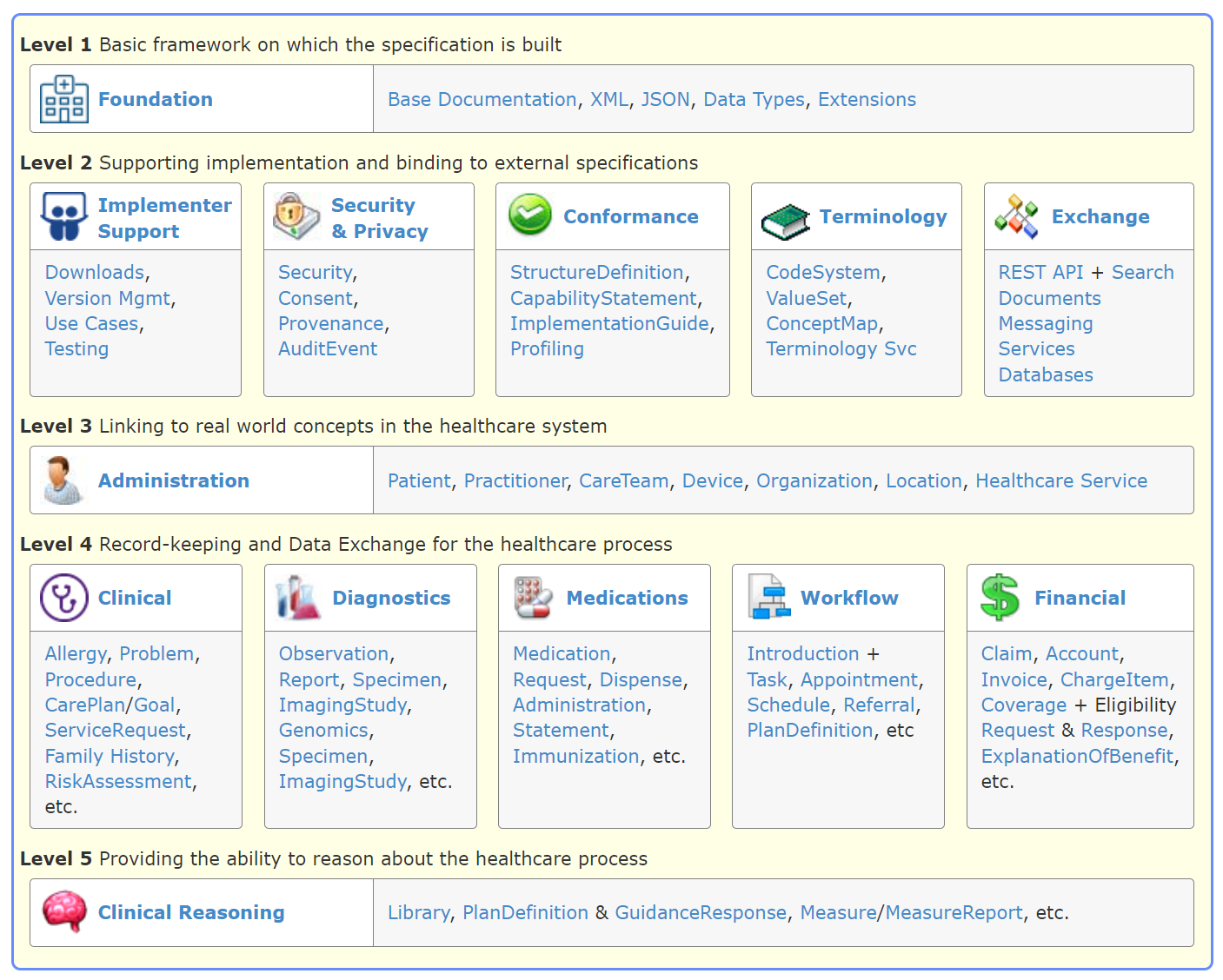
https://hl7.org/fhir/R4/index.html
OMOP CDM structure
The OMOP Common Data Model (CDM) is designed to standardize the content and structure of observational healthcare data, enhancing the efficiency of data analysis with a goal of standardizing healthcare EHR research. The model organizes data into a series of tables; among them, 17 are dedicated to clinical data while 10 are reserved for standardized vocabularies.
OMOP CDM aims to harmonize data collected for various purposes, including provider reimbursement, clinical research, and patient care. It employs a set of conventions for each table and field to ensure consistency. Data types are defined using ANSI SQL standards (VARCHAR, INTEGER, FLOAT, DATE, DATETIME, CLOB), allowing for broad compatibility and ease of use.
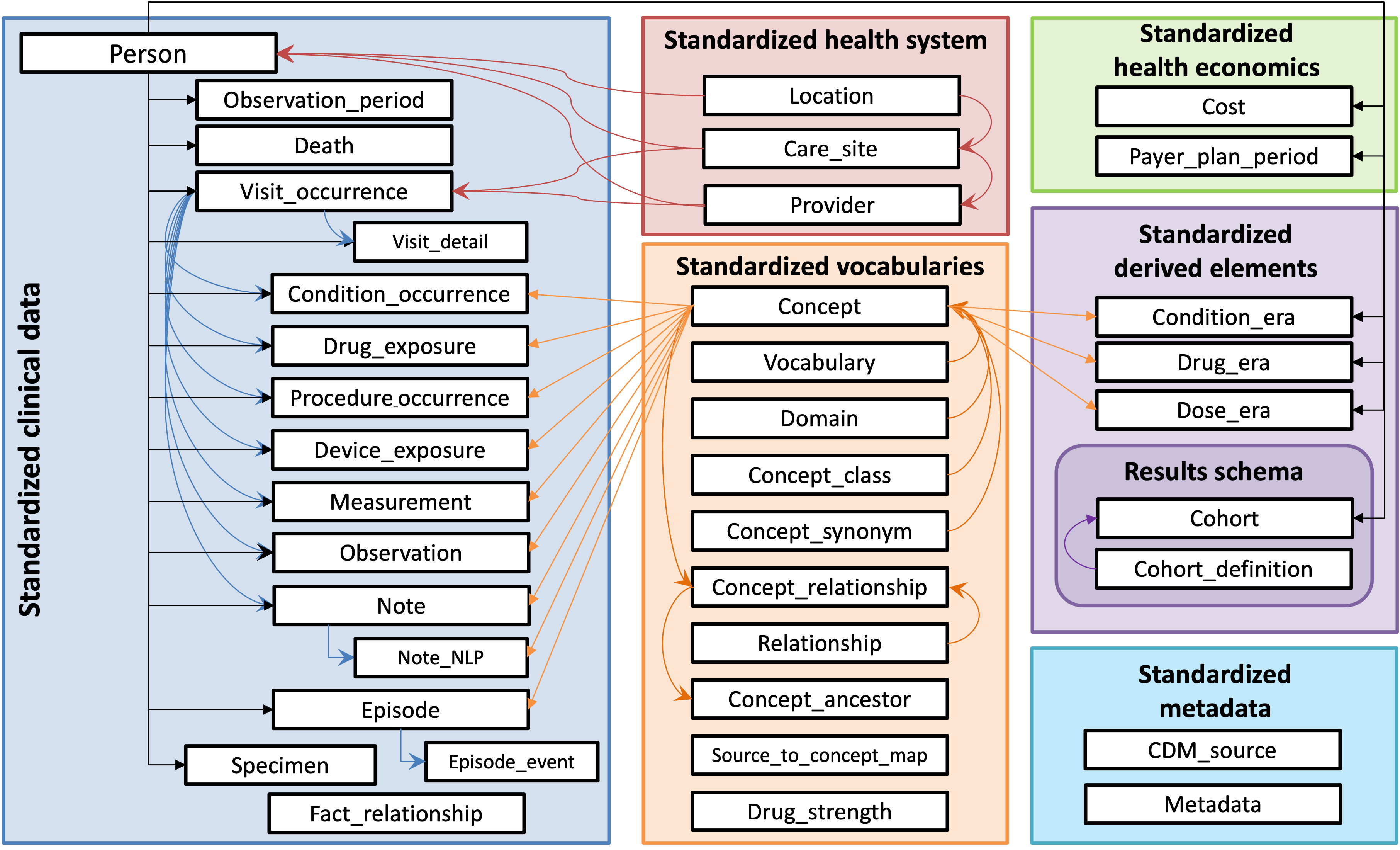
https://ohdsi.github.io/CommonDataModel/
About FHIR RDF
As a Recap, FHIR RDF is a representation format that adapts the FHIR standards to the RDF (Resource Description Framework) structure. This adaptation enhances the semantic depth of healthcare data, allowing it to be integrated with wider datasets. FHIR RDF facilitates sophisticated queries that surpass the capabilities of traditional database queries, empowering users to draw complex and semantically rich insights from interconnected healthcare information.
FHIR and OMOP CDM Comparisons
In the healthcare industry, FHIR and OMOP CDM are frequently mentioned together due to their shared focus on data consistency and interoperability. However, they differ significantly in their approaches and applications.
| Features | FHIR | FHIR RDF | OMOP CDM |
|---|---|---|---|
| Purpose and Design | Focused on real-time healthcare information exchange across various systems. Emphasizes interoperability with a flexible structure based on “resources” representing clinical concepts. | Enhances FHIR by integrating it with the Semantic Web. It represents FHIR data using RDF triples for improved data linkage and semantic richness. | Designed for standardized representation and analysis of healthcare data, especially for observational research. It aims to aggregate data from diverse sources. |
| Structure | Utilizes a resource-based structure aligning with FHIR standards. | Stores information in RDF triples format, using URIs to define resources and properties. | Adopts a table-based format, conforming to specific OMOP CDM guidelines. |
| Flexibility vs. Standardization | Offers significant flexibility, supporting extensions and customization of resources. | Provides additional semantic depth to FHIR data, enhancing interoperability and complex data integrations. | More rigid, with a fixed schema for data to fit into predetermined tables and fields. |
| Data Types and Contents | Supports a wide range of healthcare data types, including clinical, administrative, and other categories, mapped to versatile resources defined in the standard. | Encodes FHIR data in RDF, enabling intricate semantic queries and data representation. | Follows a strict guideline for data types, adhering closely to the model's structure. |
| Supported Data Formats | XML, JSON, RDF (Turtle) | RDF (Turtle), JSON-LD, N-Triples, etc. | Primarily CSV and SQL formats. |
| Interoperability | Concentrates on clinical interoperability for real-time data exchange and direct patient care. | Advances interoperability by linking with other RDF datasets and semantic web technologies. | Facilitates interoperability for research purposes, enabling data sharing and analysis across institutions. |
| Query Language | Employs RESTful APIs for CRUD operations, with queries expressed via HTTP. | Uses SPARQL for querying RDF data, supporting complex, semantic-driven queries. | Relies on SQL for querying data structured according to OMOP CDM guidelines. |
FHIR, FHIR RDF, and OMOP CDM each represent unique methods of managing healthcare data and achieving interoperability. Although invaluable to the industry, FHIR and OMOP CDM face interoperability challenges due to their differing data formats, complicating full-scale integration. This often hinders healthcare providers and researchers from fully utilizing both standards together.
However, innovations like the FHIR Ontop OMOP project demonstrate the potential for integrating these standards.
FHIR Ontop OMOP Project
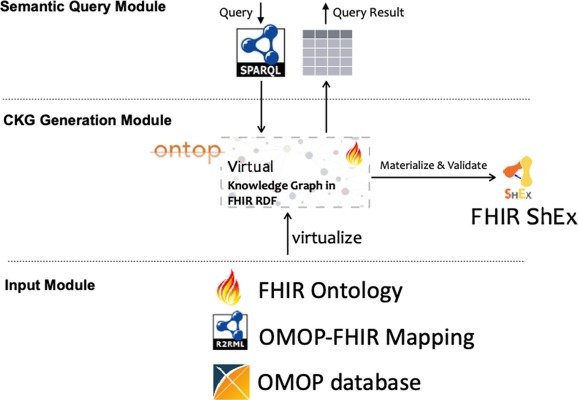
Source: ScienceDirect
The FHIR Ontop OMOP project is an important initiative that demonstrates the potential for harmonizing the FHIR and OMOP CDM standards, which are pivotal in healthcare data management. This project is designed to integrate OMOP CDM-based clinical data repositories into virtual clinical knowledge graphs, aligning with the FHIR RDF specification. It effectively allows the transformation of OMOP CDM data into FHIR RDF format, thereby enabling interoperability between these two key standards. The system utilizes virtual knowledge graph technology to facilitate this process, ensuring that the data remains faithful to the FHIR RDF model while leveraging the comprehensive data structure of OMOP CDM.
This project's success in mapping various elements of OMOP CDM to FHIR resources showcases its capability to maintain the integrity and utility of the data during transformation. The outcome is significant as it opens up possibilities for more advanced applications in healthcare, including those involving explainable AI. The FHIR Ontop OMOP project thus stands as a notable example of how interoperability between different healthcare data models can lead to more integrated and efficient data management systems, paving the way for enhanced healthcare analytics and research capabilities.
You will be able to experience it in the our system interaction - SPARQL page.For further details on the project, you can explore information available on PubMed and access the project's resources on its GitHub repository.