Introduction to Semantic Web Technologies and Electronic Healthcare Domain
FHIR
FHIR (Fast Healthcare Interoperability Resources) is a standard for electronic health care data exchange. It leverages the latest web standards with a focus on implementability.
This focus is particularly relevant given the current challenges in the healthcare industry, where managing a myriad of diverse processes often leads to significant variability in data formats. To combat these challenges, FHIR is designed to be both modular and flexible, aligning with the diverse needs of various clinical processes. It organizes data into modular components known as "Resources". These Resources can be assembled within different systems to effectively address real-world clinical and administrative issues. The modular data framework of FHIR plays a critical role in facilitating the exchange of information in ways that are both system-compatible and human-readable, thus meeting the varied demands of clinical processes.
Additionally, the adaptability of FHIR is further highlighted by its support for various data formats, including but not limited to JSON, XML, and RDF. This multi-format capability enhances its interoperability, ensuring that FHIR can be seamlessly integrated across diverse healthcare platforms and systems.
Resources:
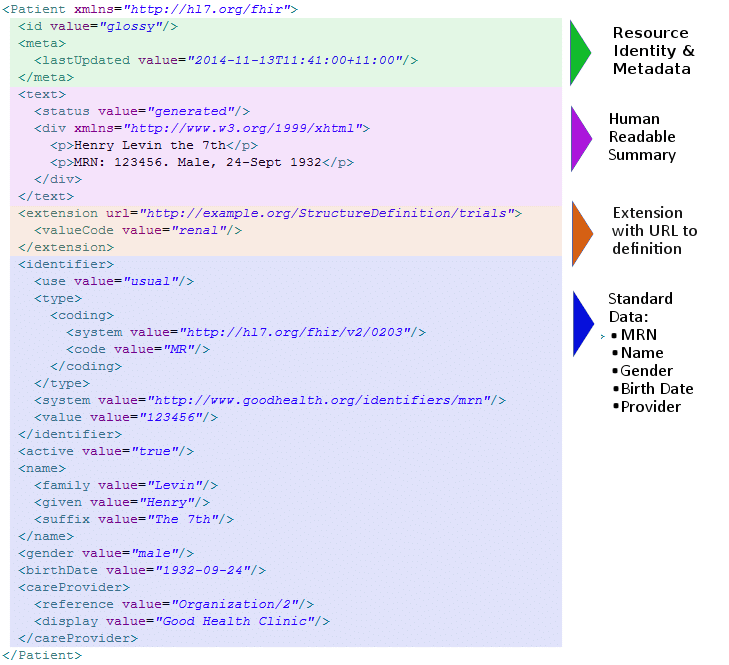
https://hl7.org/fhir/summary.html
Background on Semantic Web
In the last decade, the healthcare industry has undergone remarkable growth, largely driven by the digitization of health-related information. This evolution has sparked a need for standards capable of not only preserving information content, but also representing the intricate interconnections between data elements in a format that is both understandable to humans and readable by machines. To address this, group such as ours has been working to introduce Semantic Web technologies to healthcare. This innovation is essentially an extension of the current web, but with a crucial difference: it provides information with well-defined meaning, enhancing collaboration between computers and humans.
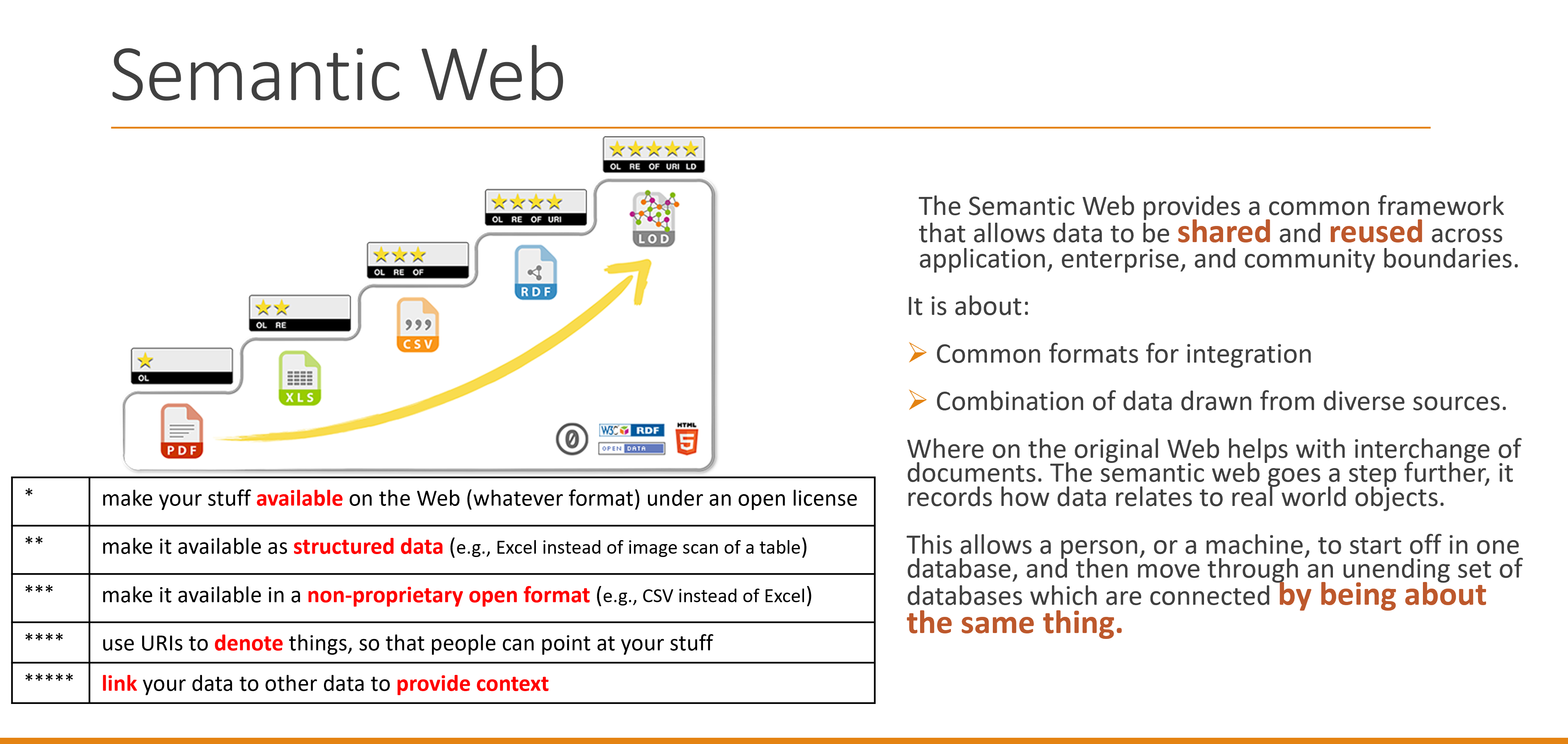
The Semantic Web stands out for its ability to provide data with meaning, thereby facilitating richer interpretations. It helps researchers to concentrate on the semantics of information, gaining deeper insights necessary for discovering disease treatments. Furthermore, this technology provides healthcare professionals with more advanced tools for personalized patient care, improving the precision of clinical management.
Further resources on topic:
- FAIR (Findable, Accessible, Interoperable, Reusable)
- The World Wide Web Consortium (W3C) - Semantic Web
Some of the technologies that are essential to Semantic Web are Resource Description Framework (RDF), Web Ontology Language (OWL), and SPARQL, which will be introduced later.
Introduction to RDF and Knowledge Graph
RDF and Knowledge Graph
RDF (Resource Description Framework) is a cornerstone technology for structured data representation and exchange on the web. In the RDF framework, every entity is considered a resource. This encompasses a wide array of objects that can be described using RDF, including web pages, people, physical objects, and even abstract concepts. Each resource is uniquely identified by a URI (Uniform Resource Identifier), ensuring that it is distinct from other resources.
RDF's strength lies in its versatile and standardized methodology for describing resources and their interrelationships. This is achieved through the use of subject-predicate-object triples, which effectively map out the connections between different entities. RDF is essential to the Semantic Web, where it provides context to data storage and transfer processes. By doing so, RDF enhances the web's capability to handle data in a more interconnected and meaningful way, forming the foundation of knowledge graphs that represent complex networks of information.

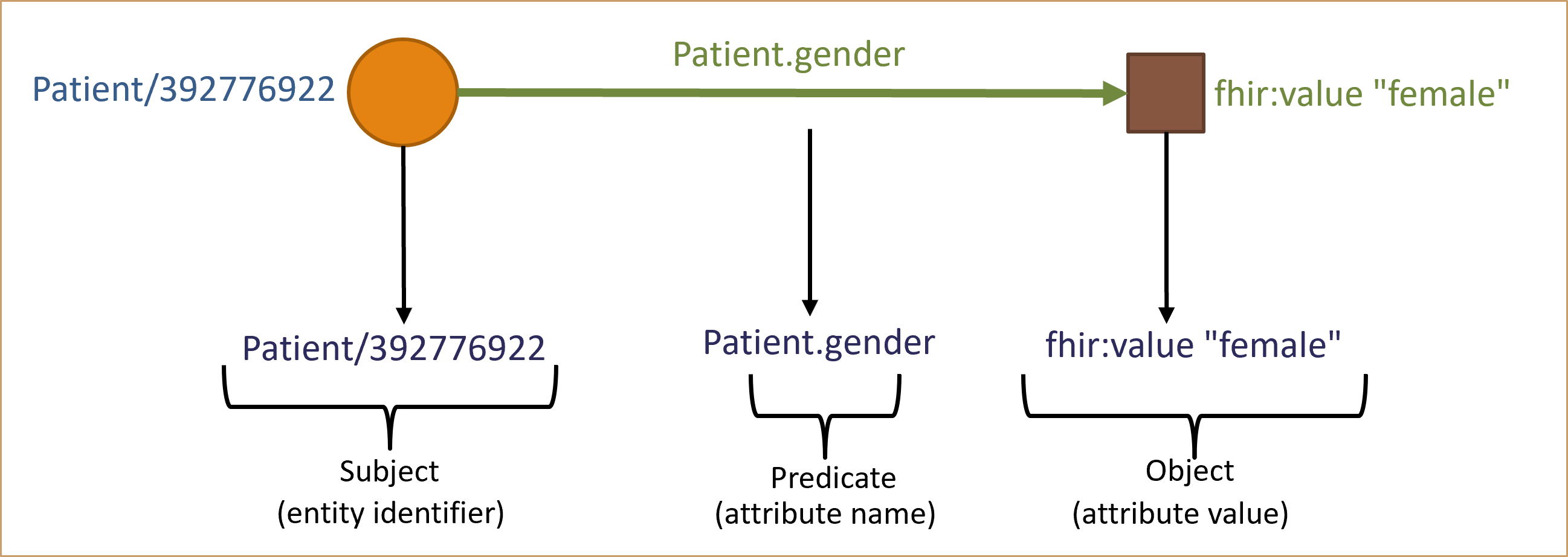
Triple Relationship
RDF extends the web's linking structure beyond mere connections between web
pages. It uses URIs to represent not just the entities but also the relationships
between them. This unique structure is commonly referred to as a "triple". The triple is composed of the three elements: subject, predicate and object.
In the triple relationship, subject represents a resource, and predicate denotes a property that
establishes a relationship between subject and object. The object in this relationship can be another
resource or a literal value, such as a string or number.
This simplistic yet powerful model allows for a seamless sharing and integration of both structured and
semi-structured data across various applications. By utilizing triples, RDF effectively enables diverse data to
be interconnected, accessed, and utilized in a more unified and coherent manner across the web.
Building Knowledge Graph From Triples
A knowledge graph represents a sophisticated method for storing and managing complex information and their interrelationships. In this structure, data is depicted in a graph format, utilizing nodes to represent entities and edges to illustrate the relationships between them. While the Semantic Web lays the theoretical groundwork and provides the technical standards for representing data on the web in a format readable by machines, knowledge graphs actualize this concept. They apply the principles of the Semantic Web to construct a structured and interconnected representation of knowledge.
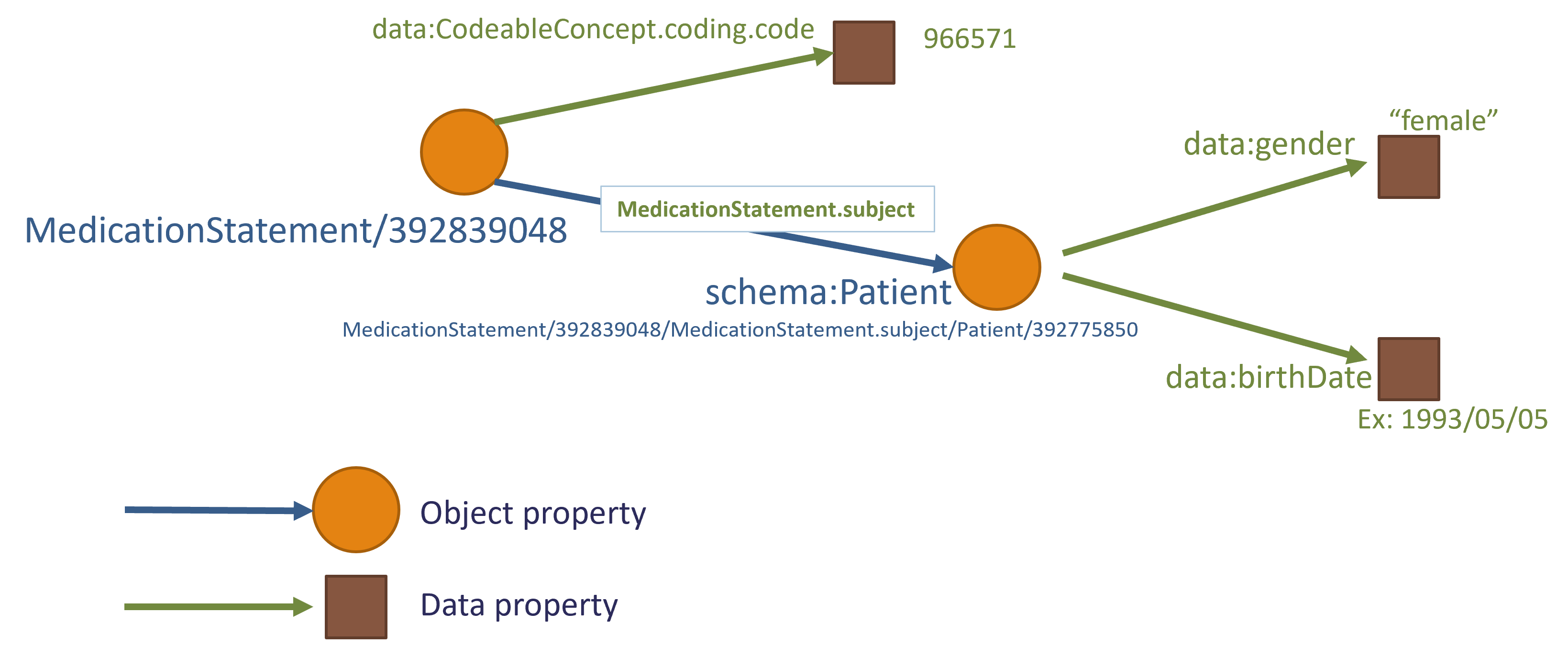
The graph as a text representation
The graph above represent the following textual relationships
| Subject | Predicate | Object |
|---|---|---|
| MedicationStatement/392839048 | CodeableConcept | (value)coding.code 966571 |
| MedicationStatement/392839048 | subject of MedicationStatement | (reference)Patient/392775850 |
| Patient/39277585 | gender | (value)female |
| Patient/39277585 | birthDate | (value)1993/05/05 |
In essence, a knowledge graph represents a specialized application or use case of RDF, where the primary focus is on representing and interconnecting structured knowledge from various sources. Knowledge graphs can be conceptualized as networks of nodes and edges. Nodes represent entities or concepts, while edges signify the relationships that exist between these entities. This structure enables a comprehensive and interconnected representation of data, facilitating efficient access and analysis of complex information networks.
FHIR RDF
FHIR RDF represents a unique data format within the FHIR framework, specifically designed to align FHIR resources with RDF formats. This alignment renders FHIR data compatible with Semantic Web technologies, significantly broadening its applications. Beyond the inherent capabilities of FHIR, the use of FHIR RDF enables healthcare professionals to execute complex queries over FHIR datasets using SPARQL, a powerful query language for RDF. This functionality not only enhances research and analytics within healthcare but also provides deeper insights from the data. Furthermore, the integration of RDF allows for the linking of FHIR resources with other data standards and ontologies, extending beyond the confines of healthcare. This cross-domain interoperability brings invaluable potentials to fields like research and public health, where correlating data from diverse sources is often crucial for comprehensive analysis and decision-making.
Turtle Format
Turtle, an acronym for Terse RDF Triple Language, is a popular representation of RDF data in an easily readable shorthand format. This format offers an efficient method for grouping URIs to form a triple, including syntactic shortcuts for abbreviating information. Turtle’s design bridges the gap between readability for humans and semantic clarity for machines, making it a valuable tool in RDF data representation.
An example FHIR RDF Observation illustrates the Turtle language and the additional
conventions used by FHIR RDF from https://build.fhir.org/rdf.html:

In this example,
- the subject's relative and absolute URIs are wrapped with '
< ... >', but when the 'fhir:' prefix or other prefixes are used, the brackets are dropped,
- the type (
fhir:Observation) and following predicates (e.g. fhir:status)
are prefixed names (like XML namespaced names),
- assertions following a '
;' reuse the same subject,
- blank (anonymous) nodes are declared with '
[ ... ]'s,
- and literals with a quoted value and an optional datatype preceded by '
^^' (e.g.
'"185"^^xsd:decimal').
SPARQL
SPARQL (SPARQL Protocol and RDF Query Language), think equivalent of SQL for RDF data. Its primary function is to enable users to extract specific data patterns from RDF datasets. SPARQL facilitates not only the retrieval of targeted data but also allows for the application of various filters to refine results, perform aggregations, and navigate the complex relationships within RDF graphs. A key feature of SPARQL queries is their structure, which consists of triple patterns mirroring RDF's fundamental triple format. These triple patterns are instrumental in formulating and expressing the structure of queries, making SPARQL an essential and powerful tool for interacting with RDF data.
Introduction to OMOP CDM and SQL
Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM)
The OMOP CDM is a standardized data model developed by the Observational Health Data Sciences and Informatics (OHDSI) program. It's designed to organize healthcare data from various sources into a common format with a consistent coding system, such as for diseases and medications. This enables researchers to conduct large-scale studies across different institutions by using standardized queries without the need to transform data for each specific study, and helps improve the efficiency in research.
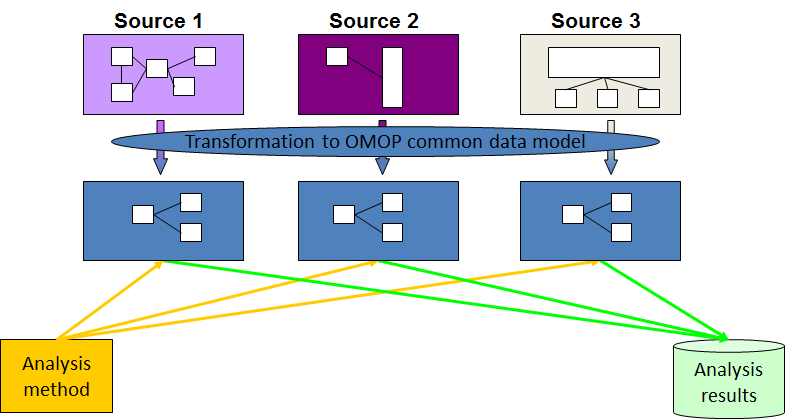
https://www.ohdsi.org/data-standardization/
Structured Query Language (SQL)
SQL is a domain-specific language used in programming and designed for managing data held in a relational database management system. It is used to insert, query, update, and delete data. With regard to OMOP CDM, SQL is the language used to interact with databases that store their data in the OMOP CDM format. This allows healthcare professionals and researchers to retrieve and analyze patient and treatment data in a uniform way, which is essential for research and decision-making in healthcare. SQL includes a variety of operations to retrieve and manipulate data, common keywords include:"SELECT", "WHERE",
"LIMIT", "ORDER BY", "GROUP BY", "HAVING", "UNION", "EXCEPT", etc.